Paul Van Liedekerke,
PhD, HDR
INRIA Paris-Rocquencourt & Sorbonne Universités UPMC Paris 6, France
tel. +33 (0)1 80 49 43 18
Paul.Van_Liedekerke@inria.fr
Current
Modeling and simulation of tissues using high resolution cell models in 3D
Mathematical models are increasingly designed to guide experiments in biology, biotechnology, as well as to assist in medical decision making. For exmaple, we may investigate the mechanical stresses experienced by the hepatocytes during liver tissue regeneration , and clarify the relation with resulting tissue architecture. Over the past couple of years, I have been elaborating on a high resolution "Deformable Cell Model" (DCM) in 3D (see Fig. 1) that is capable of representing cell shape and integrating information at subcellular scales [1,3] (PDF). In a basic DCM the cell surface is discretized by a number of nodes which are connected by viscoelastic elements, creating a flexible scaffolding structure with arbitrary degrees of freedom per cell. The model can used to simulate in vitro single cell experiments in order to extract information at subcellular scales. The triangulated structure further allows cells to interact with arbitrarily shaped structural elements. We have extended the model with the capability for the cells to grow and divide so that realistic tissue simulations are possible. I am currently using this model to investigate early embryonic development. This work is in collaboration with prof. Frédéric Lemaigre, UCL, Belgium.
The relation between cell growth and mechanical stress
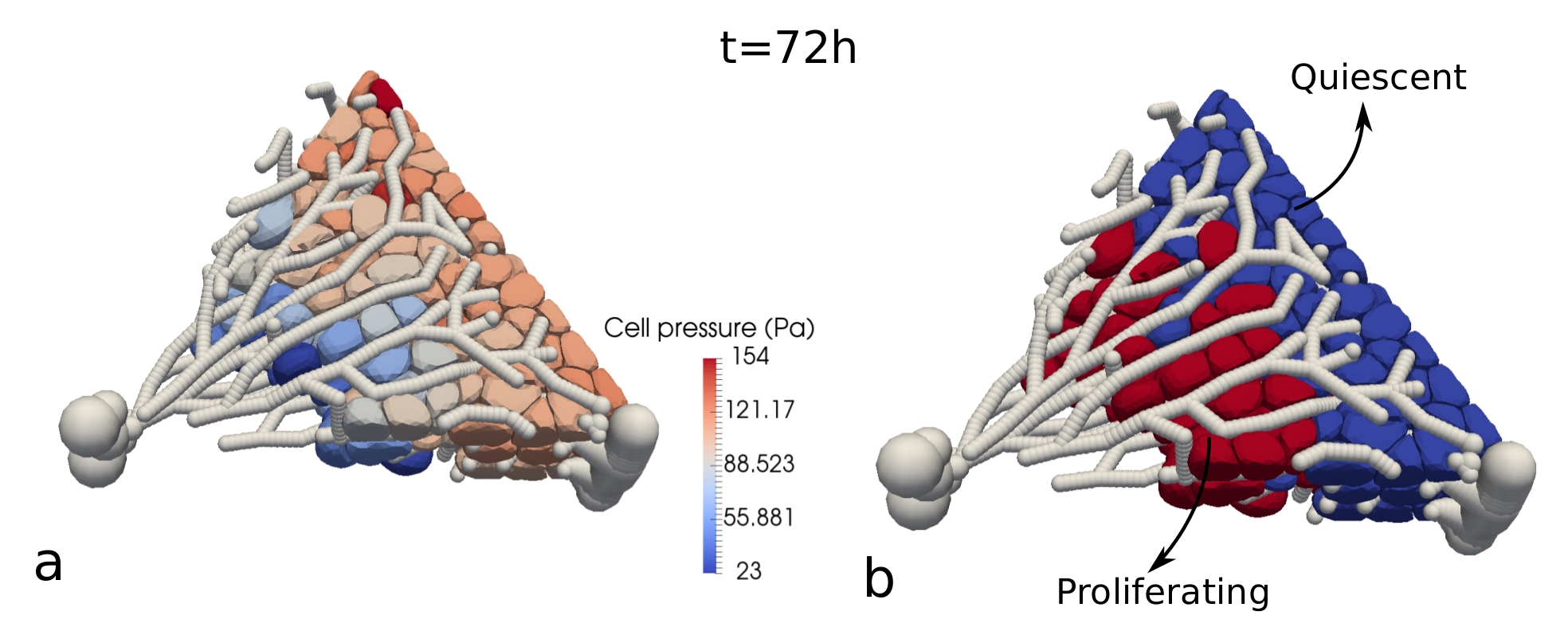
This work aims to clarify the bio-mechanical effects of growing tumor cells in confining surrounding stroma. Using agent-based models, I studied growth dynamics of tumor cells (proliferation, growth, necrosis) under different mechanical pressure conditions. One multi-cellular spheroid is growing in an elastic capsule, the other one in a Dextran solution (see Fig. 2). To simulate hypotheses concerning cell growth under pressure conditions, the model quantitatively takes into account the applied mechanical stress, cell density, and growth rate. In this work, it was demonstrated that the cellular growth response on external mechanical stress may be surprisingly quantitatively predictable over various cell lines, independently of the environmental conditions [2].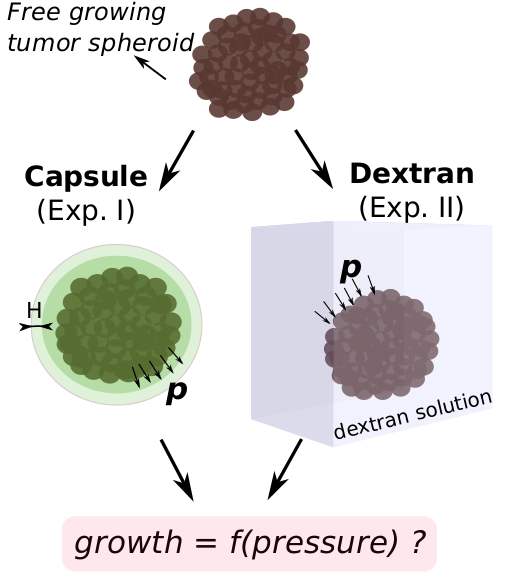
Modeling of cell-ECM interactions
The influence of mechanical feedback loop between cells and Extra-Cellular Matrix (ECM) , the medium in which cells anchor, is poorly understood. In collaboration with Dr. Andreas Buttenschoen (University of British Colombia), Dr. Margriet Palm (U. of Leiden) and Tommy Heck (KULeuven) we are creating models for cells embedded in ECM material. The cells have anchoring points (filopodia) to the ECM. The ECM is either modeled by a continuum material or a discrete network of viscoelastic elements. By a sequence of pulling and retracting the anchors, the cells move through the ECM. As such we can quantify both the stresses that work on the cells and the ECM. Part of this work is published in [4].
Past (before 2013)
Development of high resolution ”Deformable Cell Model“: modeling of arbitrary shaped cells with high detail
The majority of agent based models in the past were build on the assumption of a rigid shaped cells (so-called center based models: e.g. spheres, ellipsoids). In a some problems, such simplifications suffice to accurately simulate the biological or clinical problem (e.g. tumor growth). Yet, as more and more detail about cell shape and bio-mechanical forces is getting required nowadays, the development of more complex models has become timely.
Modeling of red blood cells in Stokes flow
Smoothed Particle Hydrodynamics (SPH) is a Lagrangian meshfree particle based method to simulate Navier-Stokes dynamics involving free surfaces, large deformations and complex physics. However, processes at cellular scale are usually viscosity dominated (overdamped) and hence a Stokes regime can be assumed. We developed an SPH solver for fluids in Stokes regime. The method (called NSPH) is based on the standard SPH technique but reduces it to a first order system and involves a different numerical solving scheme [5]. Thanks to this, longer timescales can be simulated, typically needed in cellular processes. I applied this method to the dynamics of a red blood cell passing a through a narrow channel (capillary). The blood plasma was modeled by NSPH, while the red blood cell was represented by a connection of elastic elements (see Fig.4).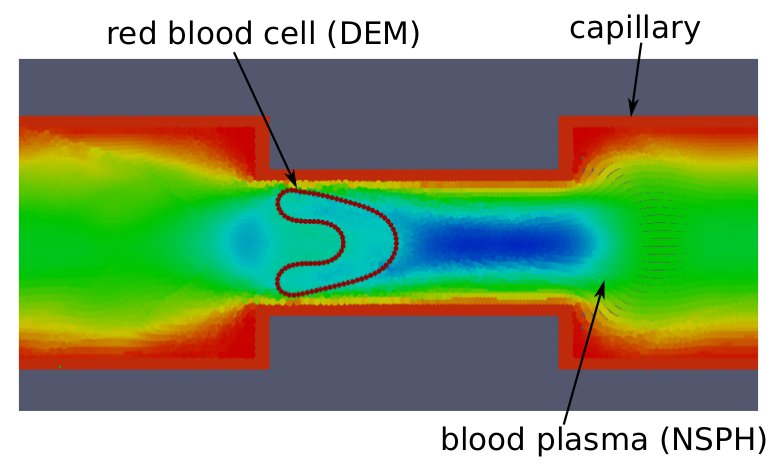
Modeling of viscoelastic bio-fluids
This project was a bilateral collaboration between KULeuven and the company MOBA N.V (The Netherlands) to understand and optimize the yield of albumen in industrial egg breaking machines. This is a very complicated process as the egg contents contain non- Newtonian fluids, membranes and complex geometries. I implemented a Smoothed Particle Hydrodynamics model for simulating viscoelastic fluids (Maxwell/Giesekus constitutive models) including surface tension. The final model encompassed a 3D realistic egg shell geometry with inside a distinct egg yolk and albumen.Modeling of impact of cellular tissues and multi-scale modeling of cellular mechanics in visco-elastic tissues.
During my late PhD and early post-doc years, I gained interest in simulation of living matter, i.e cells and tissue. I became involved in a multiscale project and collaboration between two KUL reseach groups. The goal in this project was to understand how cellular tissue responds to (dynamic) mechanical load. Hereby, the project aimed at providing insights on how two numerical methodologies in mechanical modeling of tissue, whereby one is interested in linking microscopic properties (cellular level) to macroscopic (tissue-level) behavior, can be cast into an integrative approach. This work was a scientifically very successful collaboration between the Mebios lab (Faculty of Bio-engineering, KULeuven) and the department of applied mathematics (Faculty of Engineering, KULeuven). The tissue-level is assumed to behave viscoelastic and simulated using nonlinear Finite Element Methods, while the cellular scale is approached by particle based methods and incorporated the subcellular features such as cytoplasm viscosity, cell shape, etc.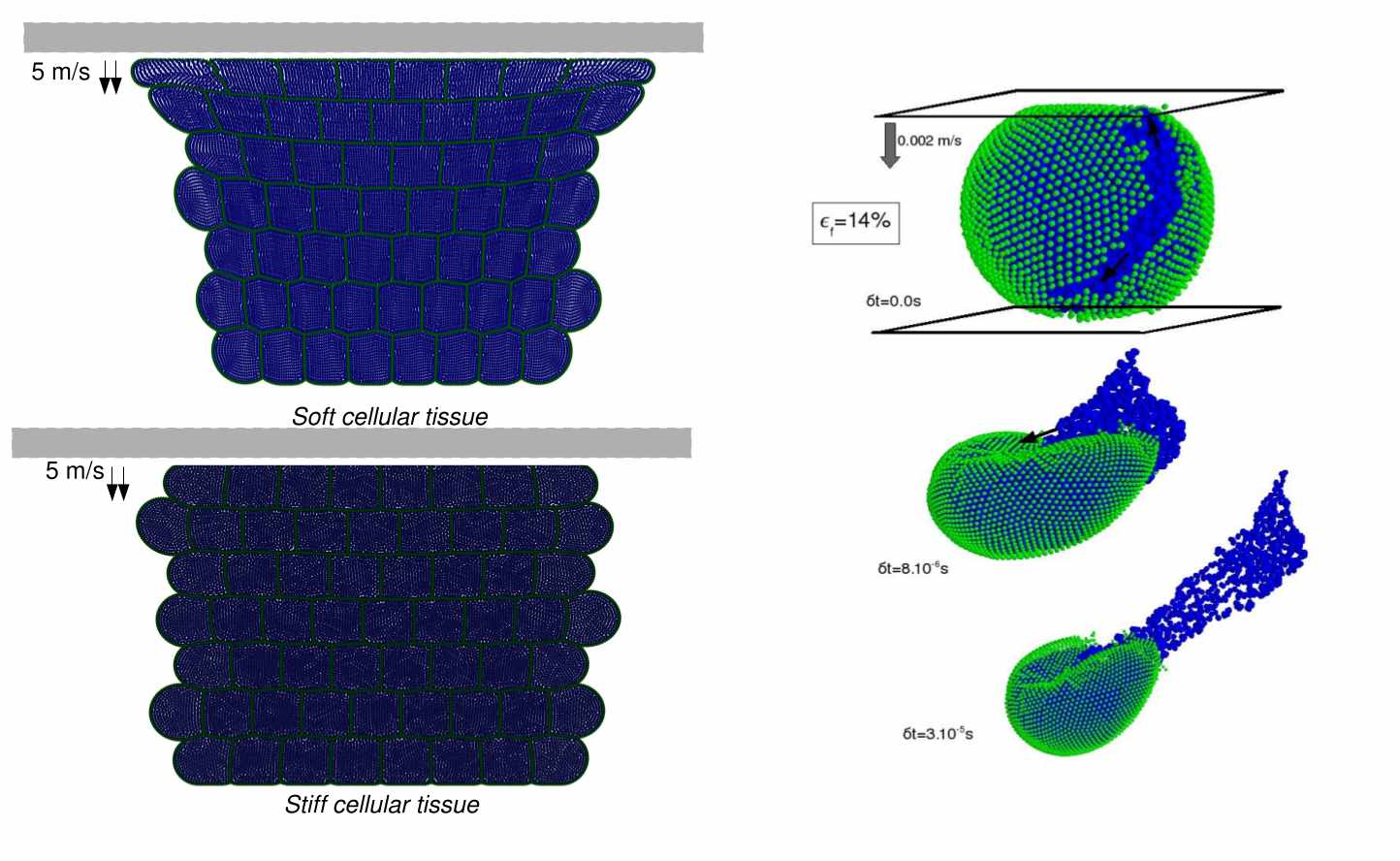
PhD work: modeling of granular material flow using the Discrete Element Method
Discrete Element Method (DEM) simulations have become standard tools to simulate granular matter flow dynamics, because of increasing interest (pharmacy, mining industry, ..) and improved com- putational means. Performing DEM simulations requires a extensive knowledge on contact mechanics, multi-body dynamics, as well as efficient contact detection algorithms, and C++ coding. Model (DEM). During my PhD, I studied dynamics and contact mechanics of rigid materials in bulk, in particular the collective motion of grains and the interaction with rigid machine parts. This involved the development and application of a particle-based code. The platform Mpacts (http://dem-research-group.com) is now a mature C++/python oriented modular code to simulate particle dynamics. It is capable of simulating solid dynamics (grains, powders, arbitry shaped objects,...) using the Discrete Element Method, as well as com- plex fluid problems (Newtonian, non-Newtonian) using Smoothed Particle Hydrodynamics. In the early years of 2000, our the KULeuven research group was one the first groups to apply this method to industrial problems. Nowadays, the software is used by several companies.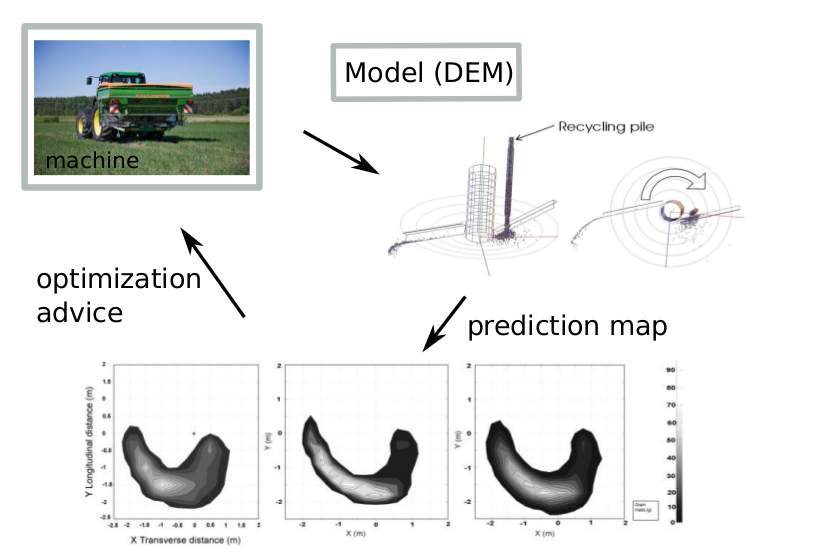
References
1. Van Liedekerke P., J. Neitsch, T. Johann, E. Warnt, S. Hoehme, S. Grosser, J. Kaes and D. Drasdo (2017) Quantifying the mechanics and growth of cells and tissues in 3D using high resolution computational models (submitted). PDF2.Van Liedekerke P., J. Neitsch, T. Johann, K. Alessandri, P. Nassoy and D. Drasdo (2017) Quantitative modeling identifies robust predictable stress response of growing CT-26 tumor spheroids under variable conditions (Plos comp. Biol, accepted).
3. T. Odenthal, B. Smeets, Van Liedekerke P., E. Tijskens, H. Ramon, H. Van Oosterwijck (2013) Contact mechanics of adhesive triangulated bodies and application to a deformable cell model. Plos Comp. Biol9(10).
4. T. Heck, S. Vanmaercke, P. Bhattacharya, T. Odenthal, H. Ramon, H Van Oosterwijck and Van Liedekerke P. (2017) Modeling ExtraCellular matrix viscoelasticity using non-inertial Smoothed Particle Hydrodynamics. Comp. Meth. Appl. Mech. Eng. 322(1).
5. Van Liedekerke P., T. Odenthal, B. Smeets, H. Ramon (2013) Solving microscopic flow problems using Stokes equations in SPH. Computer Physics Communications 184(7).
6. Van Liedekerke P., Ghysels P., Tijskens E., Samaey G., Roose D. and Ramon H. (2011) The bruising of soft cellular tissue: a particle base simulation approach. Soft Matter 7, DOI:10.1039/C0SM01261K.
7. Ghysels P., Samaey G., Tijskens B., Van Liedekerke P., Ramon H. and Roose D. (2009) Multi-scale simulation of plant tissue deformation using a model for individual cell mechanics. Phys. Biol. 6(3).
8. Ghysels P., Samaey G., Van Liedekerke P., Tijskens E., Ramon H. and Roose D. (2010) Multi-scale modeling of viscoelastic plant tissue. Int. J. Multiscale Com. Eng. 8(4).
9. Van Liedekerke P., Tijskens E., Dintwa E.,F. Rioual, J. Vangeyte and Ramon H. (2008) DEM simulations of the particle flow on a centrifugal fertilizer spreader. Powder Technology 190(3), 348-360.
10. B. Smeets,T. Odenthal, J. Keresztes, S. Vanmaercke, Van Liedekerke P., E. Tijskens, W. Saeys, H. Ramon, H. Van Oosterwijck (2014) Modeling contact interactions between triangulated rounded bodies for the discrete element method. Comp. Meth. Appl. Mech. Eng. 275(10).