(last update : 30-05-2001)
Trees of published nematode SSU sequences
(sequences available on may 2001)
Small Subunit sequences available in EMBL :
- Acanthopharynx micans [Y16911]
- Acrobeles ciliatus [AF202148]
- Acrobeles sp. [U81576]
- Acrobeloides bodenheimeri [AF202159]
- Acrobeloides bodenheimeri [AF202162]
- Acrobeloides sp. [AF034391]
- Adoncholaimus sp. [AF036642]
- Aduncospiculum halicti [U61759]
- Anisakis sp. [U81575]
- Anisakis sp. [U94365]
- Ascaris lumbricoides [U94366]
- Ascaris suum [U94367]
- Baylisascaris procyonis [U94368]
- Baylisascaris transfuga [U94369]
- Brevibucca sp. [AF202163]
- Bunonema sp. [U81582]
- Bursaphelenchus sp. [AF037369]
- Caenorhabditis briggsae [U13929]
- Caenorhabditis drosophilae [AF083025]
- Caenorhabditis elegans [X03680]
- Caenorhabditis sonorae [AF083026]
- Caenorhabditis sp. [AF083006]
- Caenorhabditis sp. [U13930] (=Caenorhabditis remanei)
- Caenorhabditis vulgaris [U13931]
- Catanema sp. [Y16912]
- Cephaloboides sp. [AF083027]
- Cephalobus cubaensis [AF202161]
- Cephalobus oryzae [AF034390]
- Cephalobus sp. [AF202158]
- Cephalobus sp. [AF202160]
- Cervidellus alutus [AF202152]
- Choriorhabditis dudichi [AF083012]
- Chromadoropsis vivipara [AF047891]
- Contracaecum multipapillatum [U94370]
- Crustorhabditis scanica [AF083014]
- Cruzia americana [U94371]
- Cruznema tripartitum [U73449]
- Cuticularia sp. [U81583]
- Cylindrolaimus sp. [AF202149]
- Daptonema procerus [AF047889]
- Desmodora ovigera [Y16913]
- Diplogaster lethieri [AF036643]
- Diplolaimelloides meyli [AF036644]
- Diploscapter sp. [AF083009]
- Diploscapter sp. [U81586]
- Dirofilaria immitis [AF036638]
- Distolabrellus veechi [AF082999]
- Distolabrellus veechi [AF083011]
- Enoplus brevis [U88336]
- Enoplus meridionalis. [Y16914]
- Eubostrichus dianae [Y16915]
- Eubostrichus parasitiferus [Y16916]
- Eubostrichus topiarius [Y16917]
- Gnathostoma binucleatum [Z96946]
- Gnathostoma neoprocyonis [Z96947]
- Gnathostoma turgidum [Z96948]
- Goezia pelagia [U94372]
- Halicephalobus gingivalis [AF202156]
- Heterakis sp. [AF083003]
- Heterocheilus tunicatus [U94373]
- Heterorhabditis bacteriophora [U70628]
- Heterorhabditis hepialus [AF083004]
- Hysterothylacium fortalezae [U94374]
- Hysterothylacium pelagicum [U94375]
- Hysterothylacium reliquens [U94376]
- Iheringascaris inquies [U94377]
- Laxus cosmopolitus [Y16918]
- Laxus oneistus [Y16919]
- Leptonemella sp. [Y16920]
- Litomosoides sigmodontis [AF227233] (sequentie is van 3' naar 5' in databank !!)
- Meloidogyne arenaria [U42342]
- Meloidogyne incognita [U81578]
- Mermis nigrescens [AF036641]
- Mesorhabditis anisomorpha [AF083013]
- Mesorhabditis sp. [U73452]
- Mesorhabditis spiculigera [AF083016]
- Myolaimus sp. [U81585]
- Oscheius dolichuroides [AF082998]
- Oscheius insectivora [AF083019]
- Oscheius sp. [AF082994]
- Oscheius sp. [AF082995
]
- Oscheius sp. [U81587]
- Otostrongylus sp. [U81589]
- Panagrellus redivivus [AF083007]
- Panagrobelus stammeri [AF202153]
- Panagrolaimoid nematode [U81580] (=Rhabditophanes sp.)
- Panagrolaimus sp. [U81579]
- Paracanthonchus caecus [AF047888]
- Parafilaroides sp. [U81590]
- Parascaris equorum [U94378]
- Parasitorhabditis sp. [AF083028]
- Paraspidodera sp. [AF083005]
- Pellioditis marina [AF083021]
- Pellioditis mediterranea [AF083020]
- Pellioditis typica [U13933]
- Pelodera punctata [AF083018]
- Pelodera strongyloides [U13932]
- Pelodera teres [AF083002]
- Philonema sp. [U81574]
- Plectonchus sp. [AF202154]
- Plectus acuminatus [AF037628]
- Plectus sp. [U61761] (=Plectus minimus)
- Poikilolaimus oxycerca [AF083023]
- Poikilolaimus regenfussi [AF083022]
- Pontonema vulgare [AF047890]
- Porrocaecum depressum [U94379]
- Pratylenchoides magnicauda [AF202157]
- Pristionchus lheritieri [AF036640]
- Pristionchus pacificus [AF083010]
- Pristionchus pacificus [U81584]
- Protorhabditis sp. [AF083001]
- Protorhabditis sp. [AF083024]
- Pseudacrobeles variabilis [AF202150]
- Pseudoterranova decipiens [U94380]
- Rhabditella axei [U13934]
- Rhabditella sp. [AF083000]
- Rhabditis blumi [U13935]
- Rhabditis myriophila [U13936]
- Rhabditis sp. [AF083008]
- Rhabditoides inermiformis [AF083017]
- Rhabditoides inermis [AF082996]
- Rhabditoides regina [AF082997]
- Rhabditophanes sp. [AF202151]
- Robbea hypermnestra [Y16921]
- Seleborca complexa [U81577]
- Stilbonema majum [Y16922]
- Strongyloides ratti [U81581]
- Strongyloides stercoralis [M84229]
- Subanguina radicicola [AF202164]
- Teratorhabditis palmarum [U13937]
- Teratorhabditis synpapillata [AF083015]
- Terranova caballeroi [U94381]
- Toxascaris leonina [U94383]
- Toxocara canis [U94382]
- Trichinella spiralis [U60231]
- Trichuris muris [AF036637]
- Turbatrix aceti [AF202165]
- Tylocephalus auriculatus [AF202155]
- Unidentified nematode [U73446] (=Rhabditidae sp. PS1010)
- Wuchereria bancrofti [AF227234]
- Xyzzors sp. [Y16923]
- Zeldia punctata [U61760]
Bootstrap distance tree :
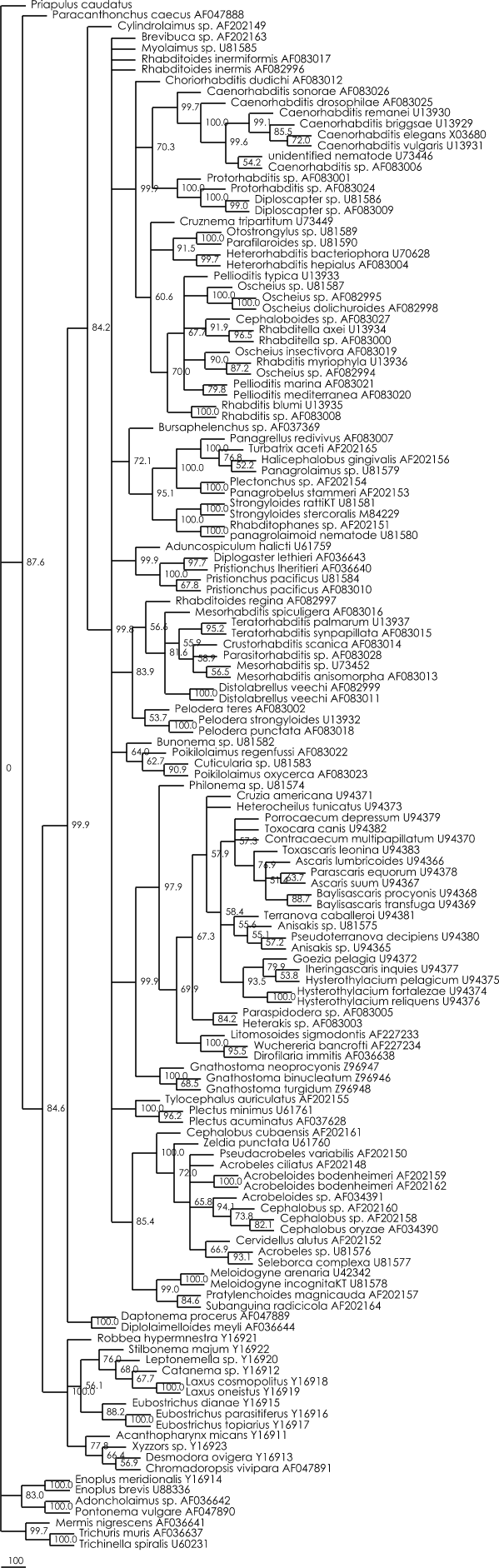
Figure 1 : Bootstrap distance tree calculated with Paup 4.0b8. (1000 bootstrap replicates, Distance measure = GTR+G+I (determined with Modeltest 3.06)).
Nexus file of this distance tree (can be viewed with Treeview)
Heuristic maximum likelihood tree :
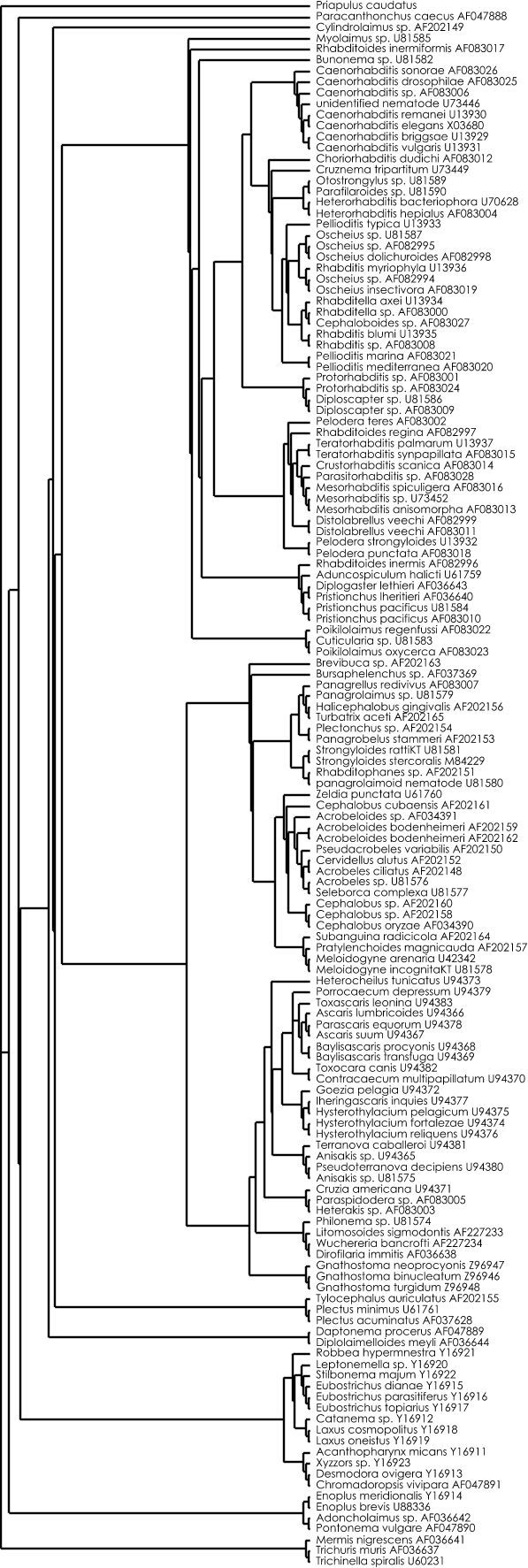
Figure 2 : Heuristic maximum likelihood tree calculated with Paup 4.0b8. (heuristic search, ML settings = GTR+G+I (determined with Modeltest 3.06), starting tree = NJ, TBR branch-swapping limitted to 24 hours (= 1900 rearrangements)).
Nexus file of this ML tree (can be viewed with Treeview)
Bootstrap maximum likelihood tree :
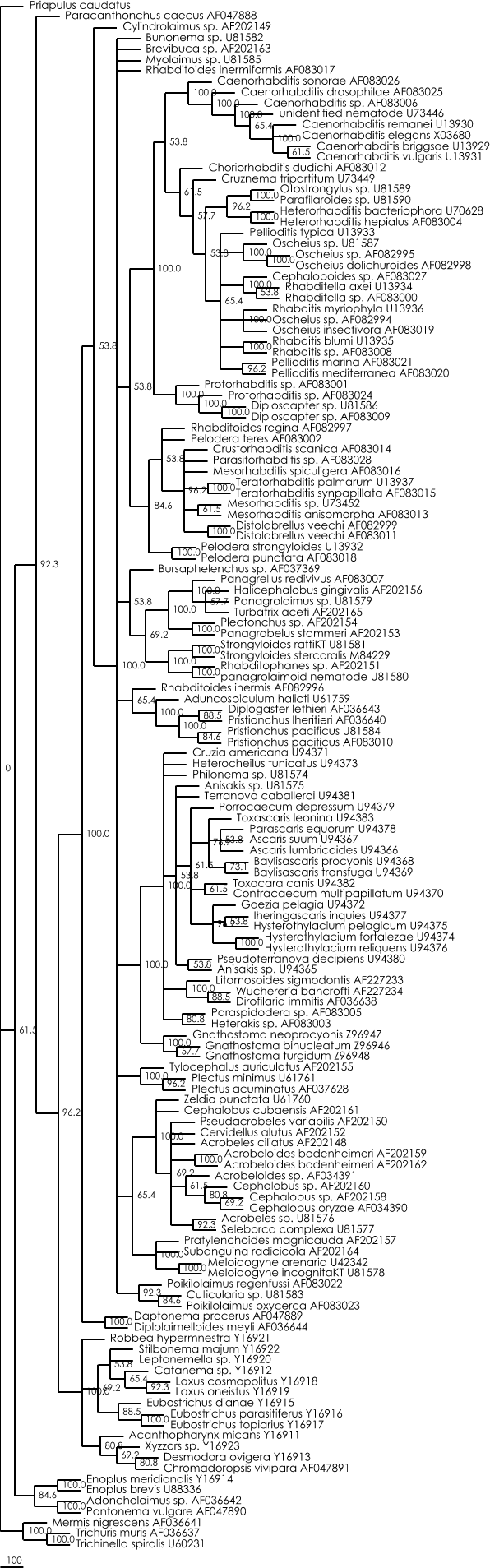
Figure 3 : Strict consensus maximum likelihood tree calculated with Paup 4.0b8. (25 bootstrap replicates, ML settings = GTR+G+I (determined with Modeltest 3.06), starting tree = NJ, TBR branch-swapping limitted to 750 rearrangements).
Nexus file of this ML tree (can be viewed with Treeview)
Bootstrap maximum parsimony tree :
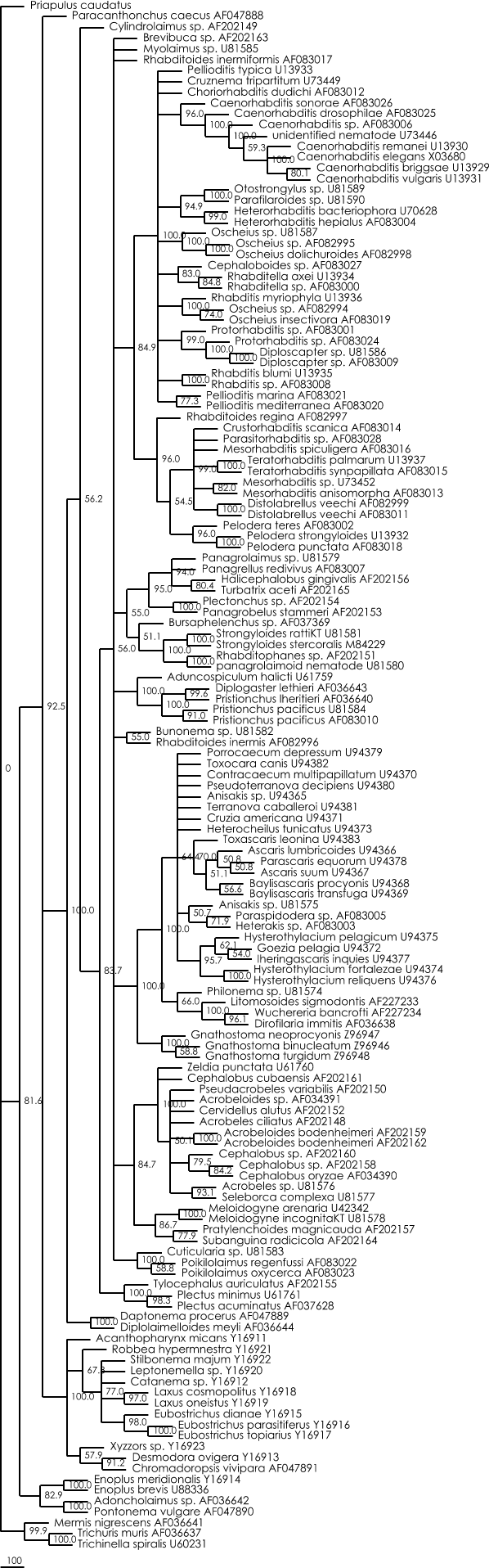
Figure 4 :Strict consensus maximum parsimony tree calculated with Paup 4.0b8. (100 bootstrap replicates, heuristic search, TBR branch-swapping).
Nexus file of this parsimony tree (can be viewed with Treeview)
back to homepage




